Exercise: RNA Transcriptionλ︎

Given a DNA strand, return its RNA complement (per RNA transcription).
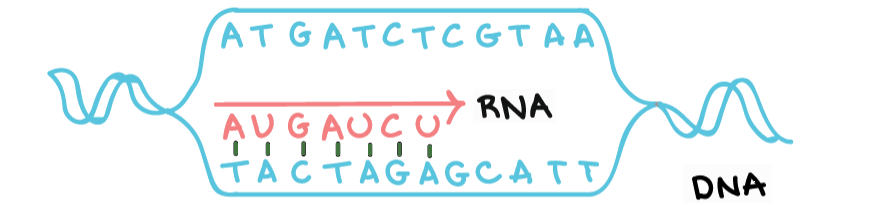
Both DNA and RNA strands are a sequence of nucleotides.
The four nucleotides found in DNA are adenine (A), cytosine (C), guanine (G) and thymine (T).
The four nucleotides found in RNA are adenine (A), cytosine (C), guanine (G) and uracil (U).
Given a DNA strand, its transcribed RNA strand is formed by replacing each nucleotide with its complement:
- G -> C
- C -> G
- T -> A
- A -> U
Code for this solution on GitHub
practicalli/exercism-clojure-guides contains the design journal and solution to this exercise and many others.
Create the projectλ︎
Download the RNA transcription exercise using the exercism CLI tool
Use the REPL workflow to explore solutions locally
Open the project in a Clojure aware editor and start a REPL, using a rich comment form to experiment with code to solve the challenge.
Designing the solutionλ︎
To convert a collection of values, define a hash-map where the keys are the initial DNA values and the hash-map values are the transformed RNA values. Using a hash-map in this way is often termed as a dictionary.
A string is used as a collection of character values by many of the functions in clojure.core. The dictionary uses characters for its keys and values.
Use the map function to pass the dictionary over the dna string (collection of characters) to create the RNA transcription.
Use an anonymous function to wrap the dictionary and pass each a character (nucleotide) from the DNA string in turn.
The result is returned as a sequence of characters.
Refactor the to-rna function and add clojure.string/join to return the RNA value as a string
(defn to-rna
[dna]
(clojure.string/join
(map (fn [nucleotide] (get {\G \C \C \G \T \A \A \U} nucleotide))
dna)))
Now the function returns a string rather than a collection of characters.
Throwing an assertion error for incorrect nucleotideλ︎
In the Exercism test suite, one test checks for an AssertionError when an incorrect nucleotide is passed as part of the DNA string.
(deftest it-validates-dna-strands
(is (thrown? AssertionError (rna-transcription/to-rna "XCGFGGTDTTAA"))))
The throw function can be use to return any of the Java errors. An assertion error would be thrown using the following code
Refactor the to-rna function to throw an assertion error if a nucleotide if found that is not part of the dictionary.
An if function could be used with a conditional to check if each nucleotide is one of the keys in the dictionary and throw an AssertionError if not found. This would mean consulting the dictionary twice, once for the conditional check and once for the conversion.
Is there a way to consult the dictionary once for each nucleotide?
The get function can return a specific not-found value when a key is not found in a map.
What if the throw function is used as the not-found value in the get function?
(defn to-rna
[dna]
(clojure.string/join
(map (fn [nucleotide ](get {\G \C \C \G \T \A \A \U} nucleotide
(throw (AssertionError. "Unknown nucleotide")) ))
dna)))
Unfortunately this approach will evaluate the throw expression regardless of if the nucleotide is found in the dictionary, so calling this version of the function always fails.
The or function evaluate the first expression and if a true value is returned then any additional expressions are skipped over.
If the first expression returns false or a falsey value, i.e. nil, then the next expression is evaluated.
Proposed Solution
Call the to-rna function with a DNA string from the unit test code
The function should return "CGAU"
Call the to-rna function with a DNA string that contains an invalid nucleotide.
An AssertionError is thrown as the X character does not exist in the dictionary hash-map, so the get expression returns nil.
Refactorλ︎
Now the function is solving unit tests, minor adjustments can be made to streamline the code.
Hash map as functionλ︎
A hash-map can be called as a function and takes a key as an argument. This acts the same as the get function, returning the value associated to a matching key, otherwise returning nil or the not-found value if specified.
(defn to-rna
[dna]
(clojure.string/join
(map (fn [nucleotide] (or ({\G \C \C \G \T \A \A \U} nucleotide)
(throw (AssertionError. "Unknown nucleotide"))))
dna)))
Anonymous functionλ︎
The anonymous function, fn, has a terse form.
#(* %1 %2) is the same as (fn [value1 value2] (+ value1 value2))
This syntax sugar is often use with map, reduce, apply functions as the behaviour tends to be compact and of single use.
If the function definition is more complex or used elsewhere in the namespace, then the defn function should be used to define shared behavior.
Solution with anonymous function
Named dictionary dataλ︎
Replace the hard-coded hash-map by defining a name for the dictionary.
Refactor the to-rna function to use the dictionary by name.
Solution using named dictionary data
(defn to-rna
[dna]
(clojure.string/join
(map #(or (dictionary-dna-rna %)
(throw (AssertionError. "Unknown nucleotide")))
dna)))
Making the function pureλ︎
Its beyond the scope of the Exercism challenge, however, its recommended to use pure functions where possible.
A pure function only uses data from its arguments.
Adding a dictionary as an argument to the to-rna function would be simple.
Pure function approach
With a dictionary as an argument the function is also more usable, as other dictionaries could be used with the function.
The function would now be called as follows